Reproducible neuroimaging
Neuroimaging tools and methods enable us to discover the underlying neuronal mechanisms of cognitive functions. Yet, we observe growing concerns about the reproducibility of neuroimaging findings. The variety of data processing pipelines applied by different labs, no code and data sharing, small sample sizes, and bad research practices undermine the trust of scientists in the results of multiple neuroimaging studies. These concerns resulted in a strong open science movement promoted by researchers around the world. In our team, we continually strive to make our research more reproducible by using open source tools, standard data organization (BIDS), and sharing our code and data. We organize and attend to Brainhack events and willingly teach students of programming, data science, and git. By collaborating closely with Student Neuroimaging Club, we seek to make neuroimaging data analysis more accessible to everyone. We develop and contribute to open-source tools dedicated to neuroimaging data processing, in particular:
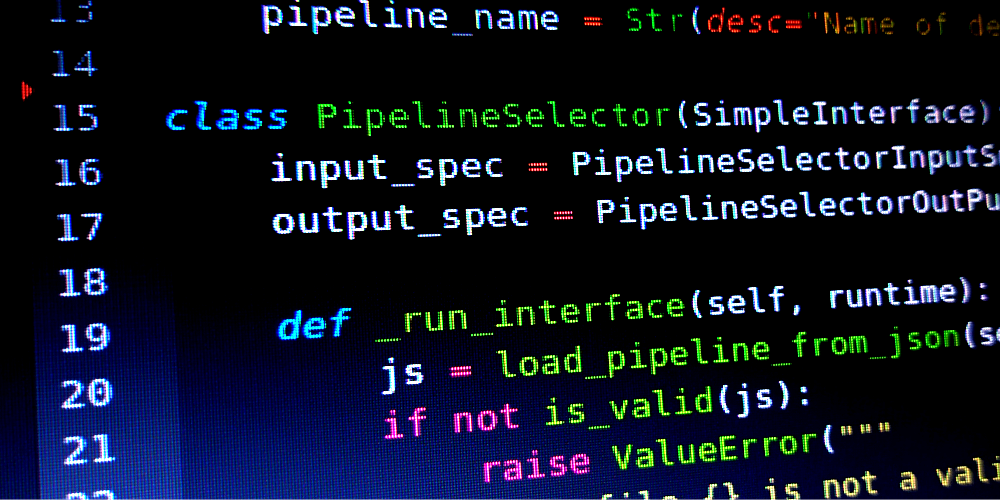
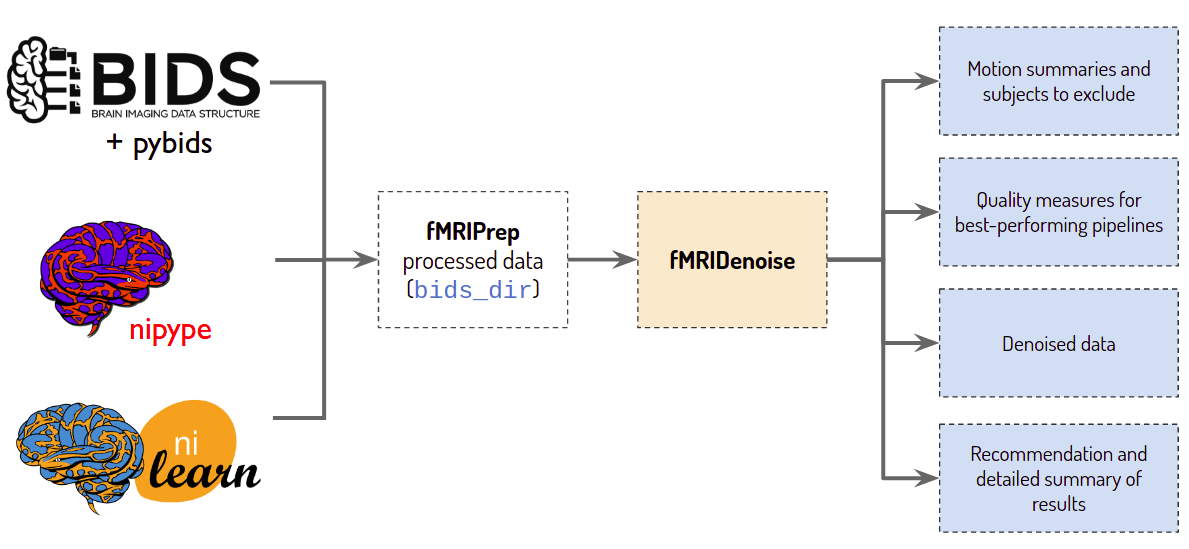
- We develop fMRIDenoise tool, dedicated to automated denoising, denoising strategies comparison, and functional connectivity data quality control
- We contribute to fMRIPrep pipeline, a robust and easy-to-use functional magnetic resonance imaging (fMRI) data preprocessing pipeline,
- We contribute to FitLins tool for estimating linear models from preprocessed fMRI data in BIDS format.
Funding:
- The Foundation for Polish Science, START & prof. Barbara Skarga fellowship,
- Ministry of Science and Higher Education, project: Excellence Initiative - Research University, awarded to Nicolaus Copernicus University in Toruń. Priority research area: Neuroinformatics.
Collaborators:
- Russ Poldrack (Stanford University, USA)
- Oscar Esteban (Stanford University, USA)
- William Thompson (Stanford University, USA)
- Chris Markiewicz (Stanford University, USA)